SOFTWARE
Information about software developed in our group.
AQUA-DUCT is a new tool facilitating analysis of the flow of solvent molecules in molecular dynamic simulations.
AQUA-DUCT allows extraction, analysis and visualization of the behaviour of solvent molecules during the entire simulation. Such analysis can be useful for the analysis of the enzymes with a buried active site, connected with surrounding solvent by tunnels. The water flow can be controlled by molecular properties of amino acids constituting tunnels or in more sophisticated enzymes by gates controlling the opening and closing of the access pathways. For more details please read our articles (AQβ, AQ1.0) or visit dedicated web page.
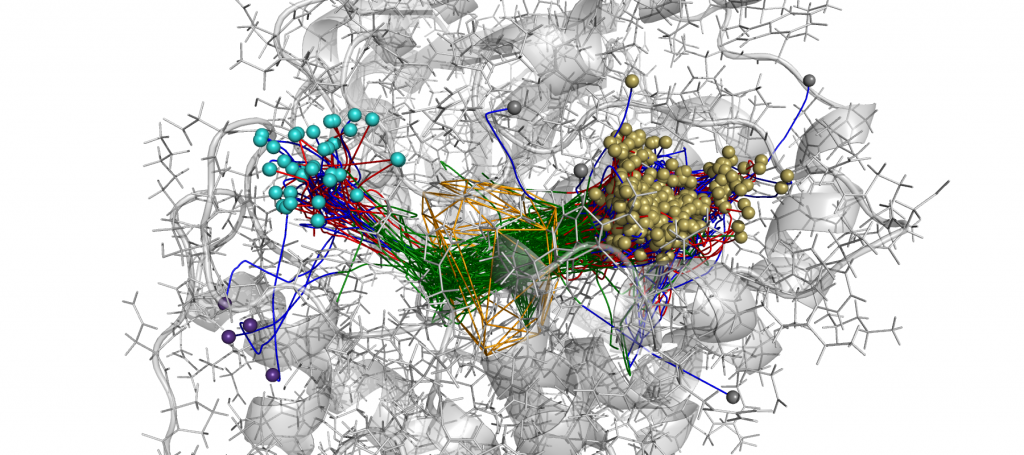
An example of AQUA-DUCT results. Approximated trajectories of all water molecules detected in active site cavity during MD simulations of M. musculus epoxide hydrolase. How to cite: 1. T.Magdziarz , K.Mitusińska, M.Bzówka , A.Raczyńska ,A.Stańczak ,M.Banas, W.Bagrowska, A.Góra, AQUA-DUCT 1.0: structural and functional analysis of macromolecules from an intramolecular voids perspective., Bioinformatics 2019, 1–3. https://doi.org/10.1093/bioinformatics/btz946 2. T. Magdziarz et al., AQUA-DUCT a ligands tracking tool. Bioinformatics 2017 btx125. doi: 10.1093/bioinformatics/btx125
PUR-GEN, a web server tailored for the automated generation of PUR fragment libraries. PUR-GEN allows users to input isocyanate and alcohol structural units, facilitating the creation of combinatorial oligomer libraries enriched with conformers and compound property tables. PUR-GEN can serve as a valuable tool for designing PUR fragments to mimic PUR structure interactions with proteins, as well as characterising simplistic PUR models. PUR-GEN
How to cite: Katarzyna Szleper, Mateusz Cebula, Oksana Kovalenko, Artur Góra, Agata Raczyńska, PUR-GEN: A Web Server for Automated Generation of Polyurethane Fragment Libraries. Computational and Structural Biotechnology Journal, 2024 doi: 10.1016/j.csbj.2024.12.004
Better ALignment CONsensus analYsis
BALCONY is an R package that facilitates the evolutionary analysis and check the variability of selected amino acids in protein structure.
One of the unique functionalities of the BALCONY package concerns the analysis of individual amino acids in primary protein structure and their neighbours in tertiary structure. Residues defining active site cavities, co-factor binding sites, selectivity filters, channels or tunnels are examples of applications for this package.For installation please go to CRAN project page: BALCONY
How to cite: A. Płuciennik, M. Stolarczyk, M. Bzówka, A. Raczyńska, T. Magdziarz, A. Góra: BALCONY: an R package for MSA and functional compartments of protein variability analysis. BMC Bioinformatics 2018, 19: 300-307..